Abstract
Background: Multiple myeloma (MM) is an incurable plasma cell (PC) malignancy typified by the presence of monoclonal proteins on serum protein electrophoresis (SPEP) and immunofixation or elevation of either kappa or lambda serum free light chains (sFLC) and end organ damage. Electronic Medical Records (EMR) offer a potential source of real world data that is more generalizable than clinical trial data and more granular than registry data. However, MM cohort derivation is challenging: diagnostic codes have poor specificity, and manually created registries are time consuming and frequently focus only on patients diagnosed on site, excluding second opinions or patients transferring care. Attempts to combine diagnostic codes with procedure codes for lab tests, biopsies or radiology studies have reported sensitivities of 73-90% and positive predictive values (PPV) of 60-93%. These approaches relied on test completion, not test results, which may misclassify patients as having MM. As part of an ongoing quality improvement project at the University of California Davis (UCD) designed to identify all patients with MM treated at our institution, we hypothesized that integrating test results, rather than their utilization, with diagnostic codes would create a more specific algorithm while maintaining sensitivity and high PPV.
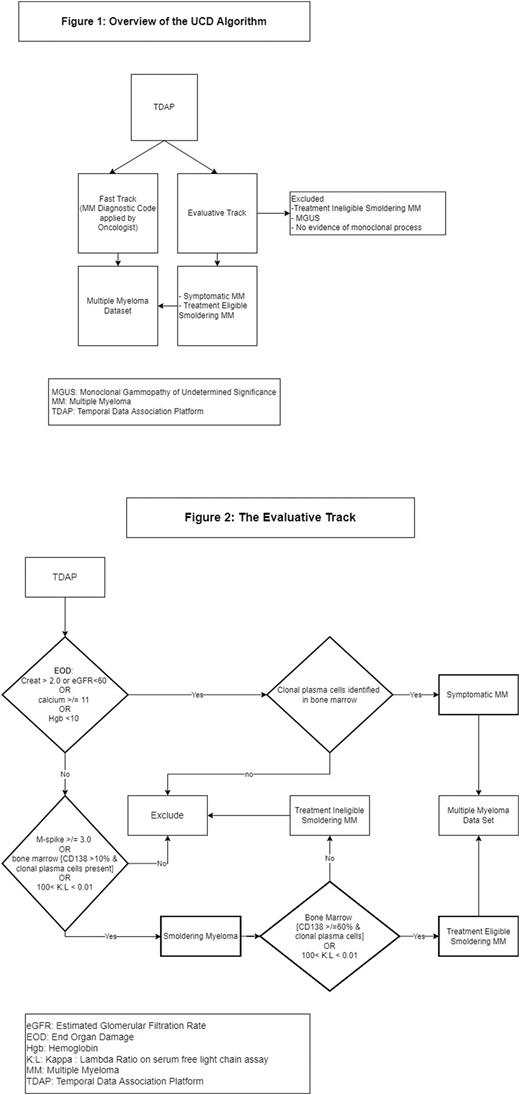
Methods: Patient diagnostic codes, lab, and bone marrow (BM) biopsy reports were loaded from EMR into the Temporal Data Association Platform (TDAP), a patient level data matrix that integrates multi-modal clinical data and organizes it by time allowing for temporal association of clinical events and the ability to assess patient trends over time. Bone marrow (BM) biopsy reports are evaluated by a machine learning algorithm trained to identify the presence and quantity (CD138 percentage) of clonal PCs. Patients identified in the screen set are then passed through both the evaluative and fast track. The UCD algorithm then runs on TDAP and consists of two components (Figure 1): an evaluative track, and a fast-track.
Like prior algorithms, the Fast-Track identifies MM specific diagnostic codes, but only evaluates visits at the UCD Cancer Center. The evaluative track classifies patients into categories based the International Myeloma Working Group diagnostic criteria (Figure 2): 1) treatment ineligible smoldering MM (SMM), 2) treatment eligible SMM, 3) symptomatic MM. Patients not meeting criteria for these categories are excluded. Patients enter the UCD MM cohort if identified as treatment eligible SMM or symptomatic MM or via the fast-track.
The UCD algorithm was built using a training set of 141 unique patients identified at the UCD Cancer Registry and sampled at random from the EMR. These charts were manually reviewed and labeled as MM, including treatment eligible SMM, (n=112) or not (n=29). To assess the performance of this and a previously described algorithm (Brandenburg 2019) once predictions were made by each algorithm, we assessed accuracy (the proportion of accurate predictions), PPV, and sensitivity.
Results: The UCD algorithm identified 111 of the MM cases and excluded MM in 9 of the negative cases with 20 false positives and only 1 false negative. This resulted in a PPV of 85% and sensitivity of 99% with an accuracy of 85%. The evaluative track alone identified 69 (sensitivity 62%) of MM cases and correctly excluded 19 negative cases, for an overall accuracy of 62%. The Brandenburg algorithm identified MM in 67 cases and excluded MM in 28 of the negative cases. It had only 1 false positive case but had 45 false negatives. This resulted in PPV of 99%, but a sensitivity of only 60% for an overall accuracy of 67%.
Conclusions: We describe a novel approach to the identification of MM patients incorporating lab and pathology results with diagnostic codes to create a sensitive algorithm while maintaining high accuracy and PPV. Evaluating test results was inadequate, and incorporating diagnostic codes allowed for higher sensitivity. Further refinement to increase specificity while maintaining sensitivity is ongoing. Performance on a validation cohort will be presented at the meeting. By incorporating test results in a temporal framework, TDAP, we can associate temporally related events such as rise in M-spike and new end organ damage. Future directions are expansion of the algorithm to other institutions and automated assessment of MM disease status.
Disclosures
Rosenberg:Takeda: Other: Institutional Research; Kangpu: Other: Institutional Research; Bristol Myers Squib: Research Funding; Adaptive: Consultancy; Janssen, Takeda: Speakers Bureau. Tuscano:Celgene: Research Funding; Genentech: Research Funding; Pharmacyclics: Research Funding; Takeda: Research Funding; Achrotech: Research Funding; ADC therapeutics: Research Funding; BMS: Research Funding. Jonas:Jazz: Consultancy, Research Funding; BMS: Consultancy, Research Funding; Gilead: Consultancy, Other: data monitoring committee , Research Funding; GlycoMimetics: Consultancy, Other: protocol steering committee , Research Funding; AbbVie: Consultancy, Other: Travel Reimbursement, Research Funding; Genentech: Consultancy, Research Funding; 47: Research Funding; Pfizer: Consultancy, Research Funding; Servier: Consultancy; Takeda: Consultancy; Tolero: Consultancy; Treadwell: Consultancy; Accelerated Medical Diagnostics: Research Funding; Amgen: Research Funding; AROG: Research Funding; BMS: Consultancy, Research Funding; Celgene: Research Funding; Daiichi Sankyo: Research Funding; F. Hoffmann-La Roche: Research Funding; Forma: Research Funding; Roche: Research Funding; Hanmi: Research Funding; Immune-Onc: Research Funding; Incyte: Research Funding; Loxo Oncology: Research Funding; LP Therapeutics: Research Funding; Pharmacyclics: Research Funding; Sigma Tau: Research Funding. Hoeg:Orca Bio: Research Funding. Kaesberg:Incyte Pharmaceuticals: Honoraria. Keegan:GRAIL: Other: Cancer Survivorship Advisory Board Meeting.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal